Other topics
Example 1 With the nested qPCR, it is possible to differentiate infections of different parasite species from same genus. Here we give step-by-step example from birds experimentally-infected with Plasmodium relictum (lineage SGS1) and Plasmodium ashfordi (lineage GRW2) (see Palinauskas et al. 2011 for details).
1) GRW2 (AF254962) and SGS1 (AF495571) sequences were retrieved from Genebank instead of sequencing.
2) Sequences were aligned with parasite 7-1 primers in Mega 5 (Remember to transfer R-primer to reverse competent for alignment)
3) Cut both ends of the sequences where the primer end. For cutting site identification, we used REsearch™ from ThermoTools However any other commercial tool or program is suitable.
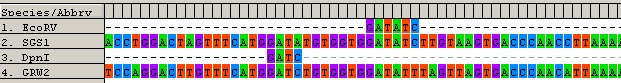
4) In SGS1, the EcoRV (GAT^ATC) cutting site was found but this was not found in GRW2. In contrast, DpnI (GA^TC) cutting site was found in GRW2 but not in SGS1 (See picture below). As a result, EcoRV can be used to digest SGS1 lineage, leaving the GRW2 lineage, whereas DpnI can be used to digest the GRW2 lineage, and leave the SGS1 lineage. After digestation, both parasites can be combined as in our standard methodology (see step 3 onwards) and the prevalence and load measured using qPCR.
Example 2
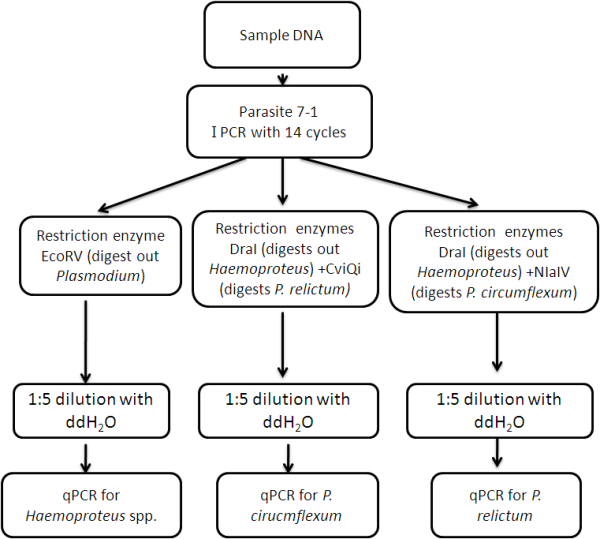
In this second example, we demonstrate how to analyse single species using a more complicated example from Cosgrove et al. (2008). In this study, they identified a number of different of Plasmodium spp. and Haemoproteus spp. parasite lineages, which can be split into morphospecies, Plasmodium relictum, Plasmodium circumflexum and Haemoproteus minutus. Sequence data for all the lineages show that the restriction enzymes EcoRV and DraI from our paper (Lipponen et al. submitted) will digest the Plasmodium spp. and Haemoproteus spp. from Cosgrove et al. (2008) . However, to resolve prevalences of the two Plasmodium spp., it is necessary to use other restriction enzymes.

Inputting sequence data into a restriction site finder e.g. REsearch™ from ThermoTools indicates that for P. relictum (SGS1, GRW11) the CviQI enzyme can be used and for P. circumflexum (TURDUS1, pBT7, pBLUTI4, pBLUTI5) the NlaIV enzyme can be used. Cosgrove et al. (2008) report a new lineage BLUTI3 that they classify as P. relictum, but shows a nucleotide blast search shows 98% similarity to P. circumflexum and only 96% with P. relictum. The species of this lineage is therefore not resolved, but has the restriction site of P. circumflexum and is digested by NlaIV.
Using these additional restriction enzyme, only a slight modification to the standard nested qPCR is needed for the nested qPCR to return results for Plasmodium circumflexum, Plasmodium relictum and Haemoproteus spp..