Normalising the amount of DNA or selecting a control gene
6. Normalisation method
Aim of this step: Normalize variation between samples
As well as identifying infected/uninfected samples, qPCR can quantify the level of parasite in the sample. This is calculated as the ratio of the amount of parasite DNA to the amount of host DNA in the sample. However, because the amount of DNA between samples will vary, data normalization must be carried out. This can be done by standardising the DNA concentration prior to usage in the qPCR, or more effectively by using a reference (control) gene. Using the DNA concentration as normalization is good method when analyzing small numbers of samples and has been used in other qPCR studies of avian blood parasites.
When normalization is done with a reference gene, host DNA specific primers are needed. Primer can be designed to anneal to any constant host gene and can be designed with free internet tools like Primer-Blast Primer-Blast or Primer3 .
7. Selecting a control gene
A key consideration when selecting a control gene is that it is found in genomic DNA, has a non variable copy number and is conserved. Ideally the fragment shold be between 80 and 250bp in length. In our study we used a 75bp fragment from the black grouse agouti-related protein (agrp) coding gene (NCBI GeneBank ID: EF571203.1). Other avian studies have used the zinc finger protein in Adelie penguins Pygoscelis adeliae or a G3PDH gene in zebra finches (Taeniopygia guttata) and Alpine swift (Apus melba) as reference genes. Here we show how to design primers for a reference gene using the rock dove (Columba livia) as an example.
The simplest method is to use an existing reference gene. We tested G3PDH gene primers previously used by Criscuolo et al. 2009 . Using a nucleotide blast query , the primers overlap and require only one base pair change (see below).
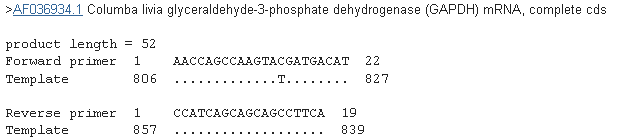
There are two further alternative. Existing primers can be tested on different species, the PCR products sequenced and the primer binding sites adjusted if necessary. Lastly, a stable non variable copy gene can be selected from Genbank if possible and primers designed with Primer-Blast . Using the sequence of the GAPDH gene in great tits (Parus major), a Primer blast query provides different primer options (see below). The next stage involves testing the primers, but in general, selecting and designing primers for a reference gene is a simple task.



