Identifying and testing restriction sites
4. Identifying the restriction sites
Aim of this step: Find parasite genus-specific restriction sites for parasite differentiation.
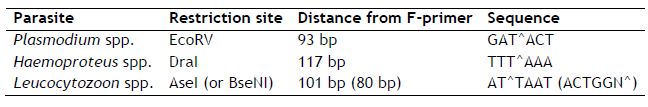
First, align all parasite sequences together with parasite 7-1 primers. Restriction sites should be between these primers. BioEdit or MEGA 4/5 are free tools for aligning the sequences. Expected restriction sites are shown in the table below. Any restriction site outside of the primers can be ignored.
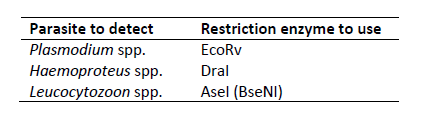
Each parasite genus should have a restriction site for one of the three (four) enzymes. For example, if you found DraI restriction site in Plasmodium spp. and Haemoproteus spp. sequences, differentiation between the genera would not be possible with these enzymes. As black grouse has an unsual Leucocytozoon spp. lineage, we had to use BseNI restriction enzyme, but AseI is most likely to be found your sample. If no restriction sites are found or they are not genus-specific, it is also possible to identify and use different restrictions sites (see here).
5. Testing of the restriction sites
Aim of this step: Test parasite differentiation with restriction enzymes and make three different restriction enzyme mixes to remove DNA of other parasites.
Take a sample that is infected with Plasmodium spp, Haemoproteus spp. or Leucocytozoon spp. and carry out a PCR with parasite 7-1 primers. Then digest the PCR-product with genus-specific enzymes and analyse the digested DNA on agarose gel.
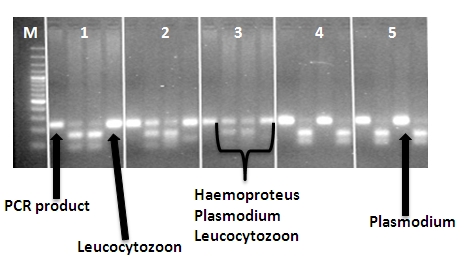
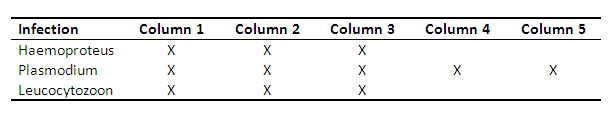
Example: The black grouse samples below are infected with differing parasites. In each column, the first band from the left is undigested PCR product, the second was digested with EcoRV + BseNI, the third with DraI + BseNI and fourth with EcoRV + DraI. If in the digested PCR products the band is at same level as the undigested PCR product, the sample is infected by parasites which the enzyme mix cannot digest.
For example: in columns 4 and 5, well 3 contains restriction fragments from DraI + BseNI restriction enzymes which cannot digest Plasmodium spp.. The undigested Plasmodium spp. can be seen as a clear band at the same level as undigested PCR product (well 1), whereas the other wells (2 and 4) containing EcoRV show only digested product.